PRESGENE SERVER
USER GUIDE
(Version: PRESGENE - v1.0.0)
The tutorial is divided into three sections.
A.
Navigating through the PRESGENE server webpage
B.
Understanding the Input Files required by the Server for the Prediction
C.
Essential Gene Prediction using PRESGENE- Step-By-Step Guide
A. Navigating through the PRESGENE server webpage
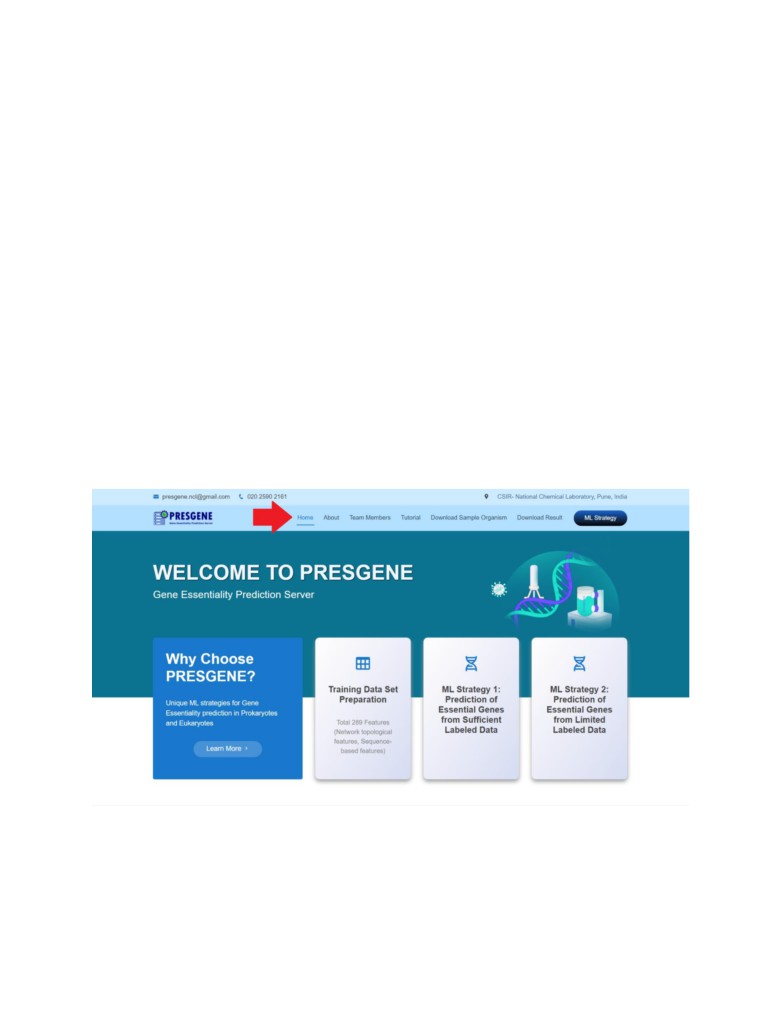
1. Home Page
Click here to know more about the PRESGENE server
2. Machine Learning Strategies Implemented
PRESGENE server offers two ML Strategies [1,2] for prediction of essential genes in prokaryotes
and eukaryotes.
➢ For information on the Supervised Learning based Strategy: Click on ML Strategy 1
➢ For information on the Semi-supervised Learning based Strategy Click on ML Strategy 2
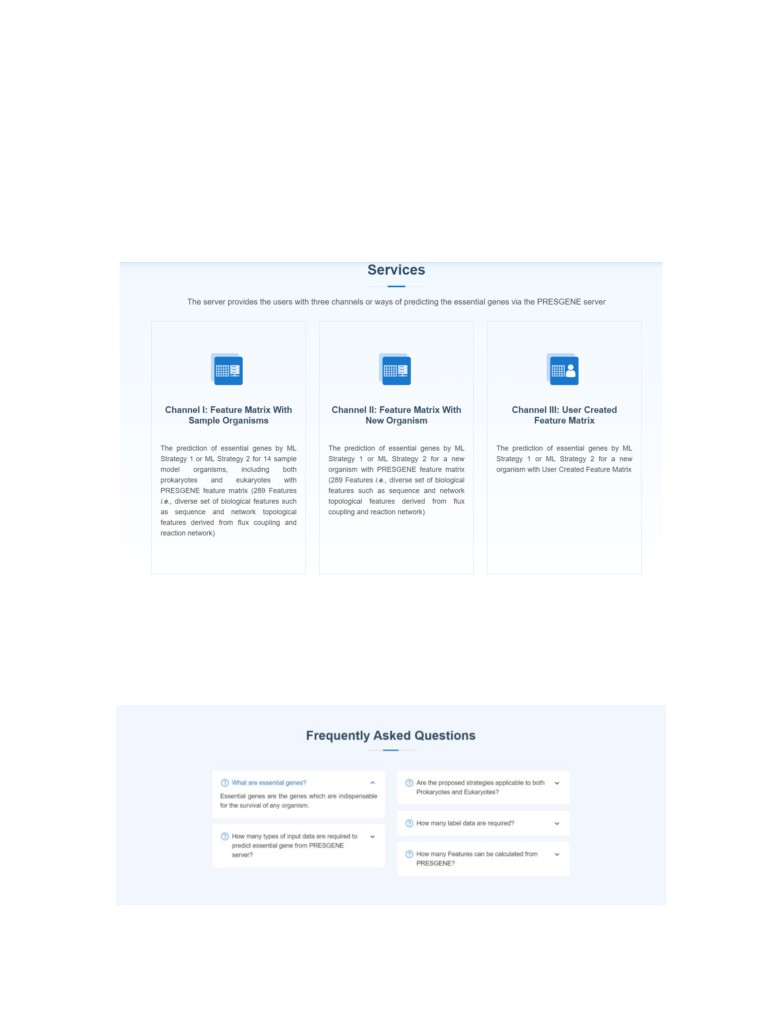
3. Services of the PRESGENE server
The server provides the users with three channels or ways of predicting the essential genes via
the PRESGENE server.
Channel I: This provides the option to the user to test the pipelines on 14 sample model
organisms, including both prokaryotes and eukaryotes. The user can choose to vary the
percentage of labeled data to be used for the prediction of the essential genes. The results
produced for these model organisms through the server can be directly incorporated by the users
in their own study for prediction of drug targets or other applications.
Channel II: This helps the user to predict essential genes for a new organism using the
PRESGENE server in four simple steps. To prepare the training dataset, the user needs to provide
the name of the organism and five input files (discussed in section B).
Channels I and II direct the user to the Dataset Preparation (Feature Matrix Calculation) tab to
calculate and predict essential genes using the ML1 or ML2 strategies using 289 biological
features.
Channel III: The server provides the user with an option to incorporate and test the influence of
other biological features (apart from the existing 289), calculated and provided to the server in the
form of a User Created Dataset with Feature Matrix through Channel III. This matrix forms the
training dataset of the pipeline and should include the various features as columns and the
reaction-gene combinations (samples) of the metabolic network as rows. The last column of the
matrix should contain the gene essentiality information as E (Essential), N (Non-Essential), or UD
(Undefined) as target variables. Channel III will directly take the user to the Training and
Prediction tab of the ML pipeline.
The step-by-step tutorial for essential genes prediction using Channels I, II and III have been
provided in Section C.
4.
Frequently Asked Questions
Hover to the Frequently Asked Questions section on the Home Page to get answers to the
common queries.
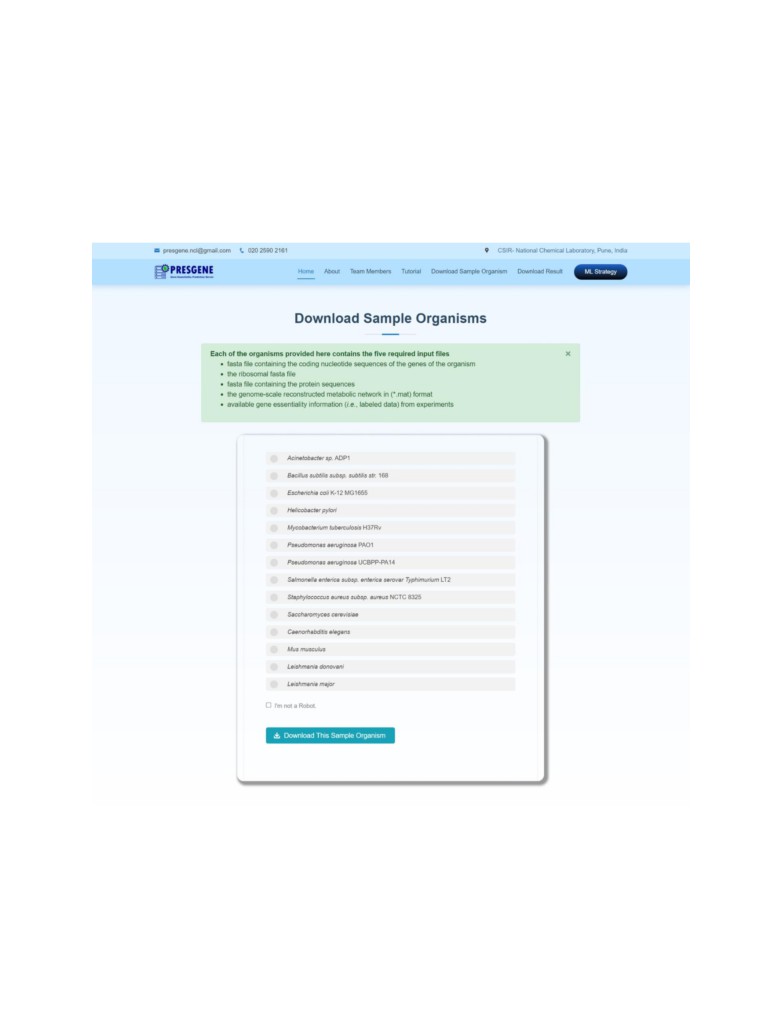
5.
Download Sample Organism
The server provides the user the input data files of 14 sample model organisms as a reference
dataset in a readily usable format that can be used to test the pipelines.
B. Understanding the Input Files required by the Server for the
Prediction
We have provided a set of sample input files (in Download Sample Organisms Tab) for the ease
of first-time users of the server to understand the file formats required as input in prediction for
Channels I or II. The five input files for each organism have been provided in .zip format.
A description of each input files has been provided below.
Input File 1: Metabolic Network File
This is a genome scale reconstructed metabolic network (GSRMN) file (*.mat). The genome-
scale reconstructed metabolic network contains the information of metabolites, reactions, and
genes. These networks are available throughout the literature and the BIGG database [3].
Input File 2: Nucleotide sequence File
This is a multi-fasta file which includes nucleotide sequences of all the protein coding genes of
your organism of interest arranged in the form of FASTA format.
Please make sure that each sequence starts with the below indicated header line which starts
with “>” followed by the Gene ID. Users can use any type of Gene ID (e.g., Refseq, NCBI, EMBL,
GenBank, DDBJ, etc) to construct their input files.
It is important that this same GeneID is maintained across all the other input files for
uniformity in collecting, calculating and assembling the features for the final features’ dataset
matrix.
Input File 3: Ribosomal Sequence File
This is again a multi-fasta file of ribosomal sequences of your organism of interest arranged
similarly in a FASTA format.
Note how the gene ID in the header line is maintained same as that of the nucleotide
sequence file.
Input File 4: Protein sequence File
This is a multi-fasta sequence file of the proteins encoded by the coding nucleotide
sequences(cds) of the genes of your organism of interest. It is essentially a sequence of amino-
acids in a FASTA format, similar to the above two files; Nucleotide sequence file and Ribosomal
sequence file
Input File 5: Gene Essentiality File
This file contains known experimental information about the essentiality of the genes of the target
organism for model training. This is the labeled dataset in .csv format which essentially contains
two columns- Gene, Class
In the column ‘Class’, mention the essentiality information of the Gene list of the target organism.
It is to be noted that a minimum of 1% labeled dataset is required for model training to perform
essential genes prediction using PRESGENE.
✓ Essential gene is to be denoted as: E
✓ Non-essentiality is to be denoted as: N
C. Essential Gene Prediction using PRESGENE- Step-By-Step Guide
User can start gene essentiality analysis by clicking ML Strategy tab.
The two broad steps in the prediction pipeline are:
1) Dataset preparation
(Feature matrix table) from the essential sequence, flux and
essentiality information of the user query organism (sample/ new organism)
2) Model Training and Prediction using supervised or semi-supervised ML Strategies.
PRESGENE provides users three options for feature matrix calculation (dataset preparation) for
gene essentiality prediction - Channel I, Channel II and Channel III.
Channel I: Can be used when user wants to test or work on any of the sample organism data
provided readily by PRESGENE
Channel II: Can be used when user wants to perform prediction on new organism for which the
user has available the five required input data files and nominal (minimum 1%) essentiality
information for the novel organism of their interest.
Channel III: Can be used if the user already has a ready feature matrix (Master Dataset) and
want to directly run. See below in Channel III section to know about the format of a ready feature
matrix file.
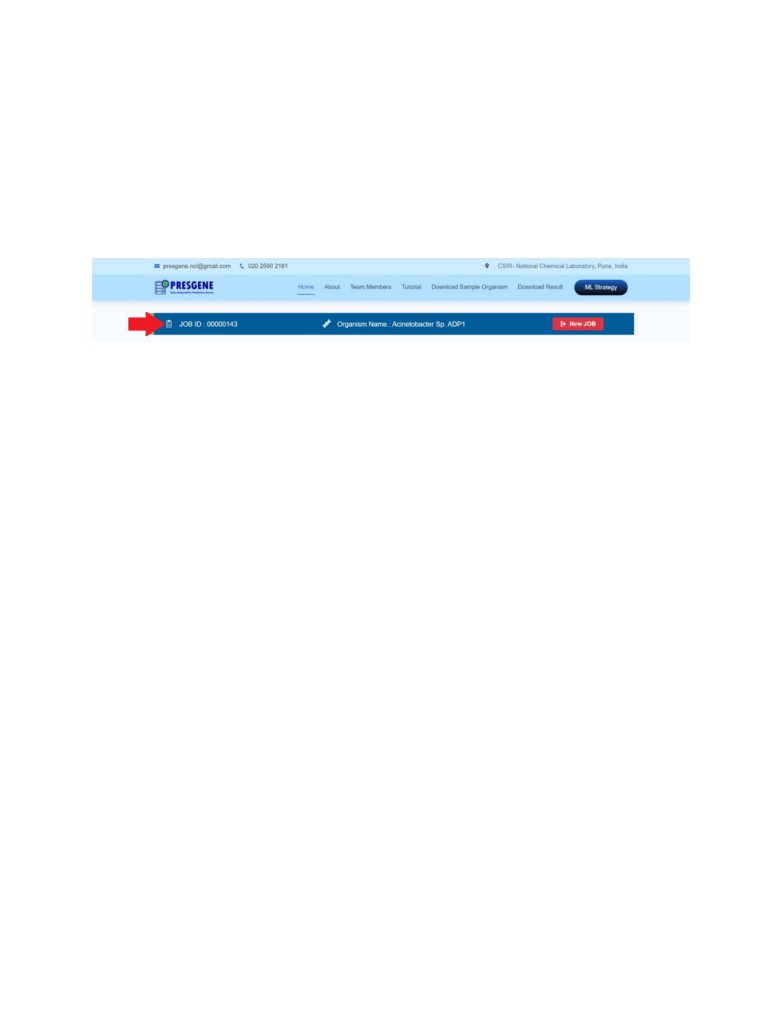
JOB ID: Each submission of the user is assigned a unique JOB ID which is an 8-digit
number such as ‘00000143’. User should note this JOB ID to track and retrieve results
which will be available in the server for next 15 days.
New JOB
By clicking on New JOB, ongoing JOB will be terminated and a new JOB ID will be assigned.
Channel I: Running PRESGENE on Sample Organisms
STEP 1: Select the organism of your choice from the list of 14 available sample organisms. Select
the precent gene essentiality information available. It is kept at default value of 30%. User can
vary it according to their choice.
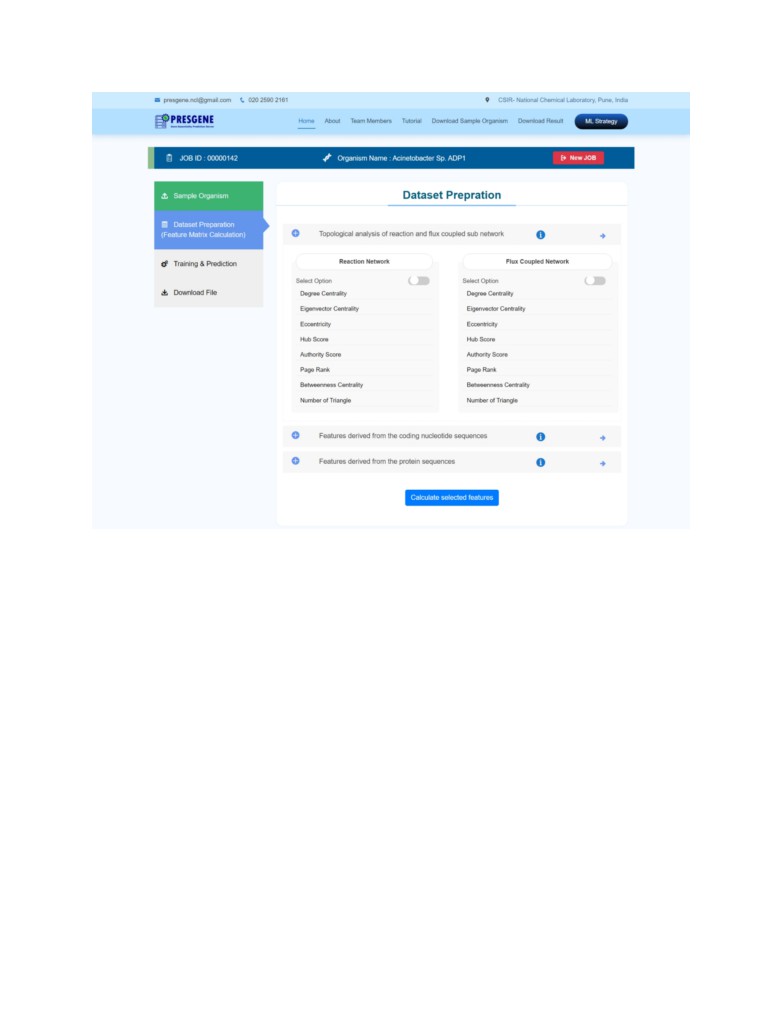
STEP 2: Next step is Dataset (Feature matrix) preparation. Click on the tab on the left side. The
Dataset Preparation tab allows the user to choose the set of biological features that the user
wishes to consider for the gene essentiality prediction. However, it is recommended to consider
all 289 biological features for higher accuracy and better prediction of essential genes. The list of
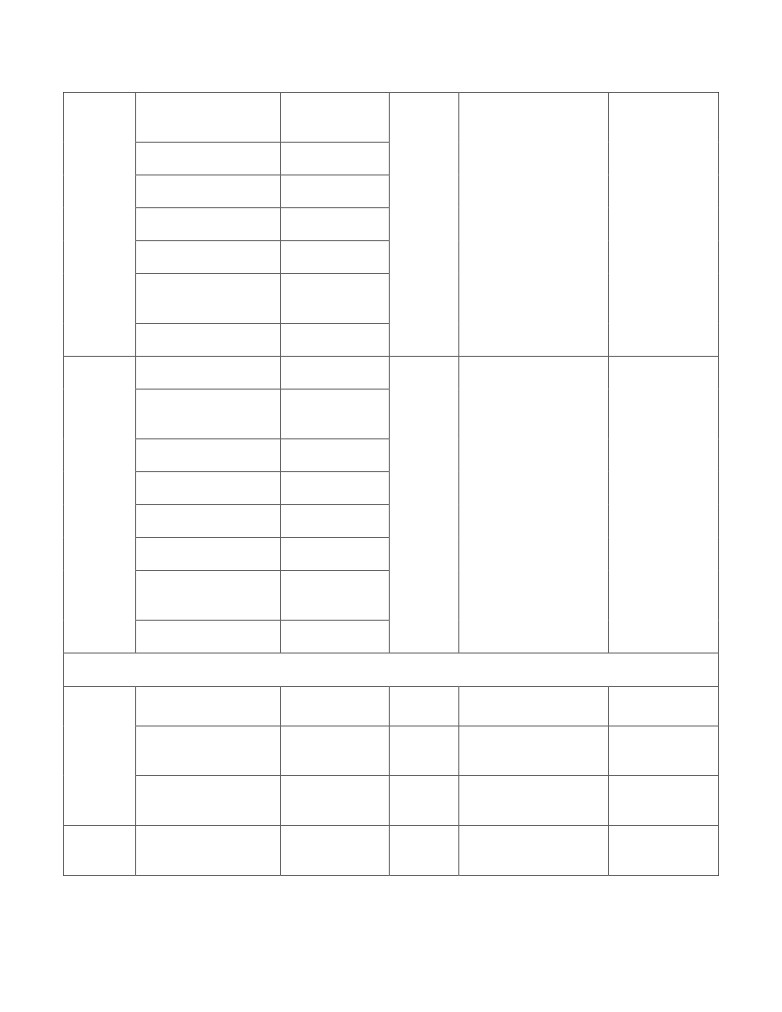
features has been enlisted in Table 1.
Table 1. List of Features and the software packages used feature calculation
Feature
Abbreviation of
# of
Programming
Features name
Software Packages
Types
features name
features
Languages
Topological analysis of reactions and flux-coupled sub-networks
Degree Centrality
TF_RN_DC
8
Eigenvector
TF_RN_EC
Centrality
Eccentricity
TF_RN_ET
The COBRA Toolbox to
generate the reaction
Hub Score
TF_RN_HS
network from Genome
scale metabolic
Reaction
MATLAB, R,
Authority Score
TF_RN_AS
network (.mat)
Network
Perl
Page Rank
TF_RN_PR
"igraph" for network
Betweenness
analysis[4]
TF_RN_BC
Centrality
Number of triangle
TF_RN_NT
Degree Centrality
TF_FC_DC
Eigenvector
TF_FC_EC
Centrality
Eccentricity
TF_FC_ET
F2C2 tool v0.95b (Flux
Couple Analysis)
Flux
Hub Score
TF_FC_HS
MATLAB, R,
Coupled
8
Perl
Network
Authority Score
TF_FC_AS
"igraph" for network
analysis[4]
Page Rank
TF_FC_PR
Betweenness
TF_FC_BC
Centrality
Number of triangle
TF_FC_NT
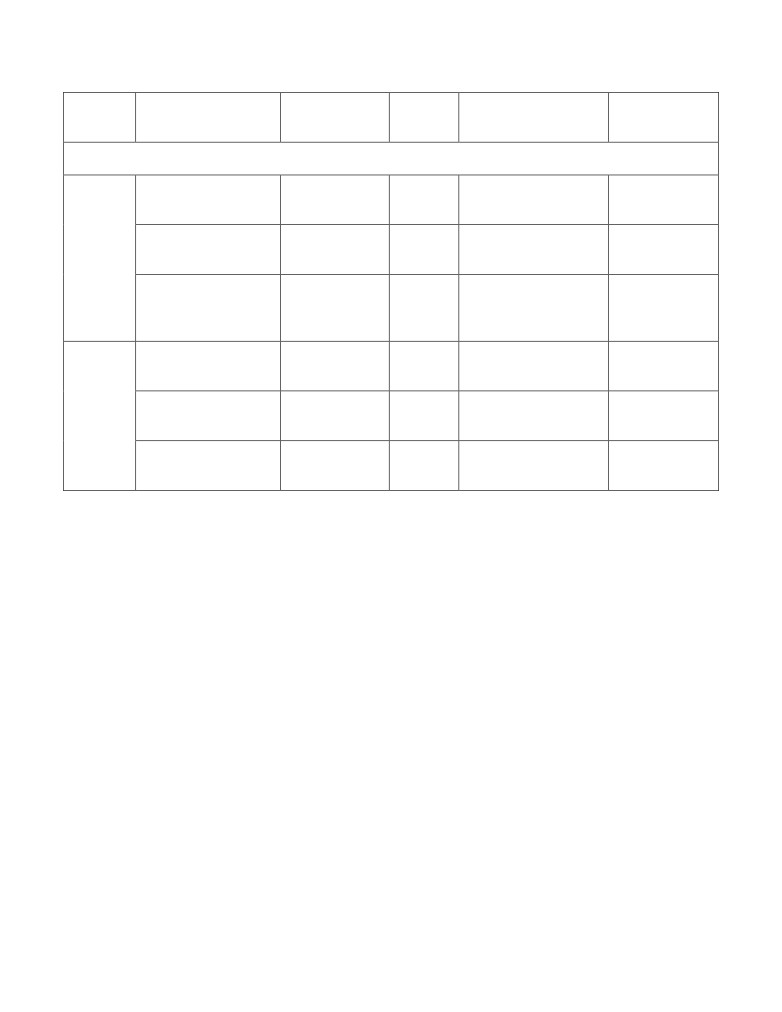
Features derived from the coding nucleotide sequences
Nucleotide content
NS_DF_NC
4
In house Perl script
Perl
Effective Number of
EMBOSS package
Derived
NS_DF_ENC
1
Perl
Codons
version 6.6.0-1[5]
features
Codon Adaptation
EMBOSS package
NS_DF_CAI
1
Perl
Index
version 6.6.0-1[5]
Mutual Information
Informati
NS_ITF_MI
16
in house Perl script
Perl
(MI)
on-
theoretic
Conditional Mutual
features
NS_ITF_CMI
64
in house Perl script
Perl
Information (CMI)
Features derived from protein sequences
Frequencies of the
EMBOSS package
PS_DF_FA
20
Perl
twenty amino acids
version 6.6.0-1[5]
EMBOSS package
Derived
Protein length
PS_DF_PL
1
Perl
version 6.6.0-1[5]
features
Paralogy based
features (Paralogy
PS_DF_PS
6
BLAST [version 2.2.26]
Perl
score)
Fourier sine
PS_ITF_FSC
70
in house Perl script.
Perl
coefficient
Informati
on-
Fourier cosine
PS_ITF_FCC
80
in house Perl script.
Perl
theoretic
coefficient
features
Average Kidera
PS_ITF_AKF
10
in house Perl script.
Perl
Factor
Click on Calculate selected features.
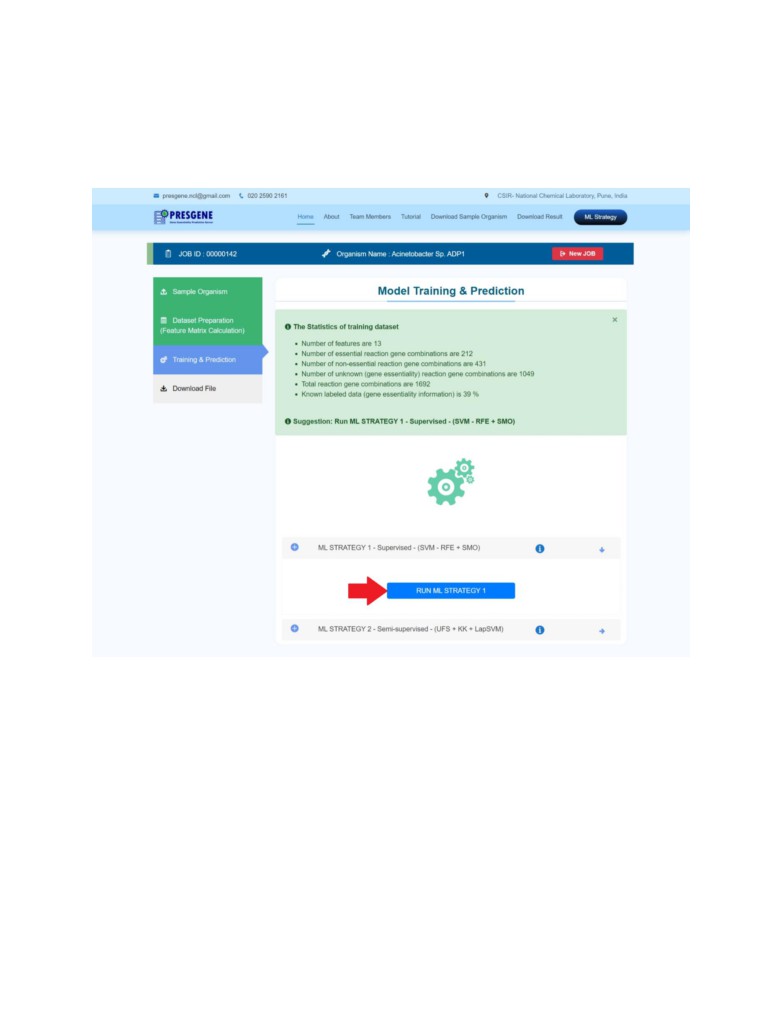
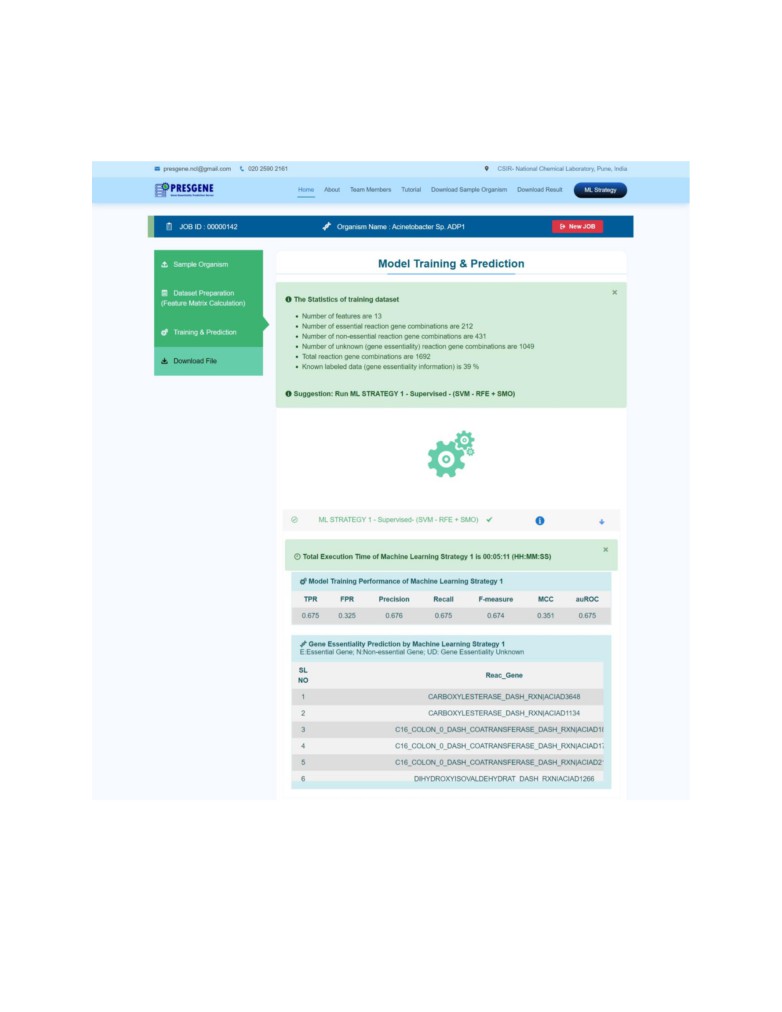
STEP 3: Next, click on the third step Training and Prediction.
PRESGENE provides the training dataset statistics. Based on the availability of the information
the user gets a suggestion to either use ML Strategy 1 or ML Strategy 2.
Please click on the suggested strategy to run it.
STEP 4: Once the prediction is completed, Expand the selected strategy to browse the results.
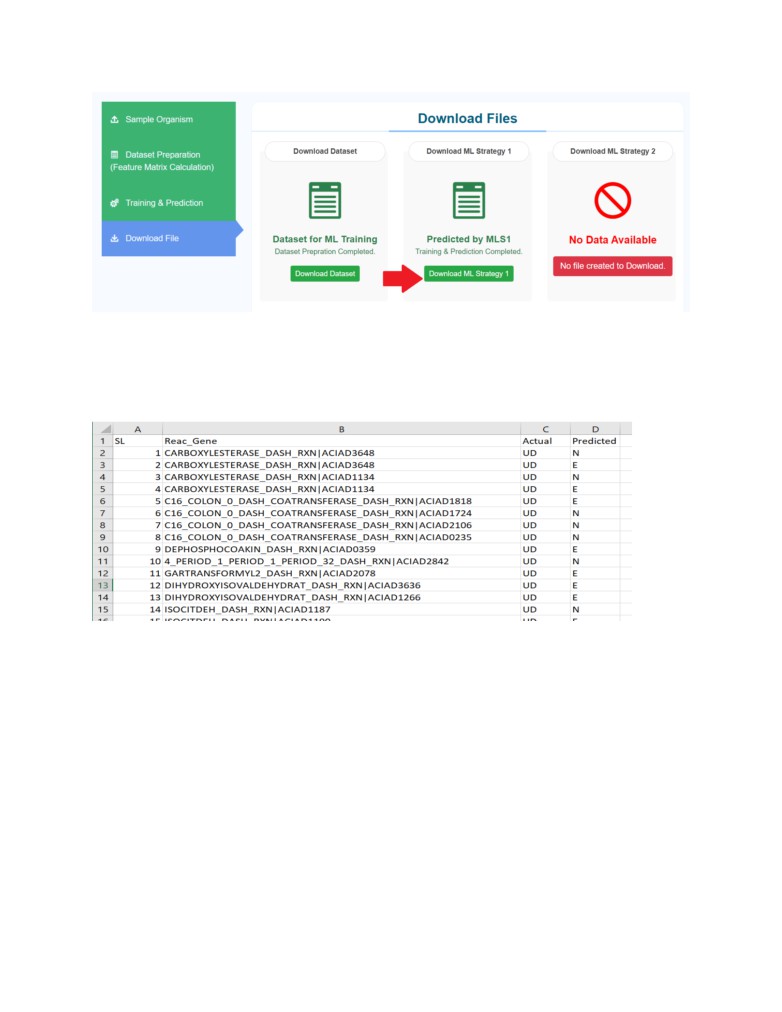
Browse to the last tab of the Pipeline Download Files to download the final prediction result
from PRESGENE.
The results are tabulated in a csv file and contains the prediction result in the form of “E” or “N”
notations corresponding to each gene where “E” indicates the gene to be predicted as “Essential”
and “N” indicates the gene to be predicted as “Non-essential”.
Channel II: Essential Gene prediction for new organism
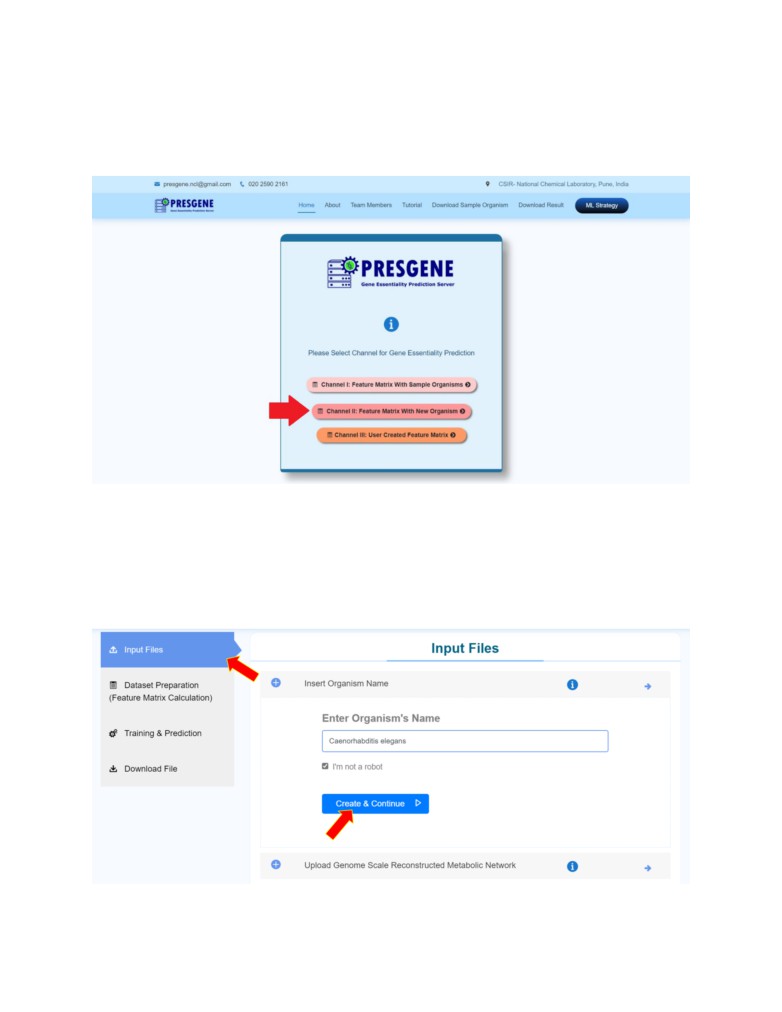
STEP 1: Click on Input Files. Enter the name of the organism on which the user wishes to predict
essential genes and click on the Create & Continue to go to the first step of pipeline Input Files.
Once the user has five input files ready for their organism of interest, please follow the following
steps to carry out essential gene prediction.
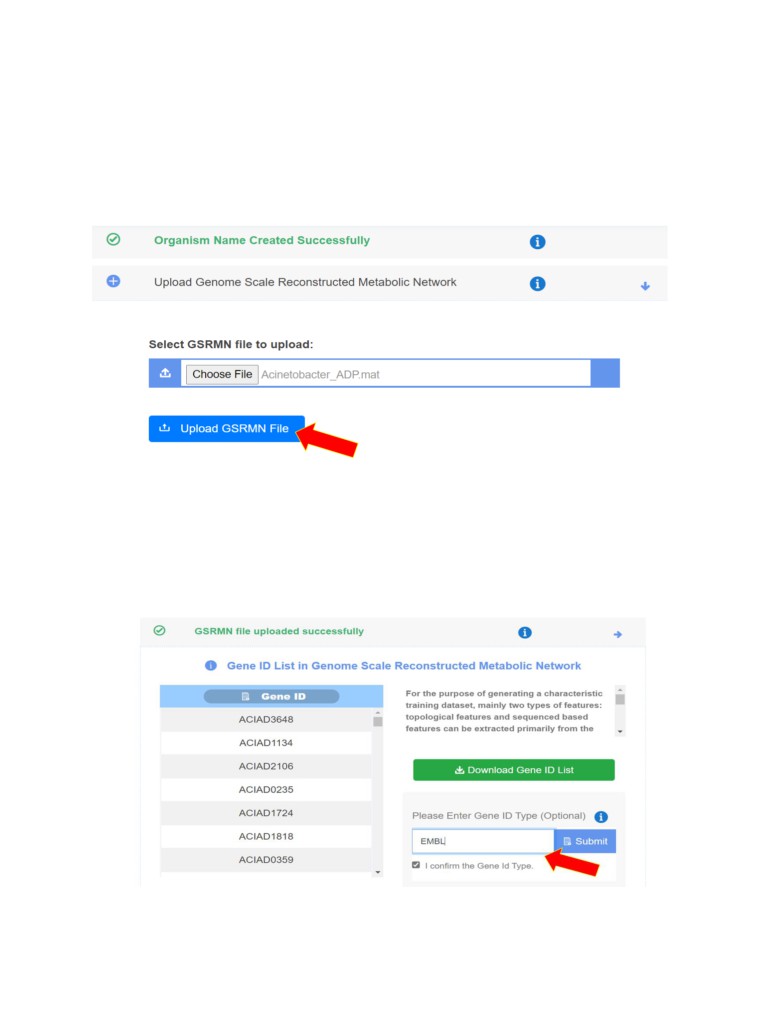
• Upload the Genome-Scale Reconstructed Metabolic Network (GSRMN) file by clicking on
the Choose File. Once the file is selected, click on Upload GSRMN File.
• Next, input the Gene ID Type and click on “I confirm the gene ID type”. This step is
optional.
• User can take a look at the Gene ID list here to confirm if all the genes have been read
by the algorithm correctly for further processing.
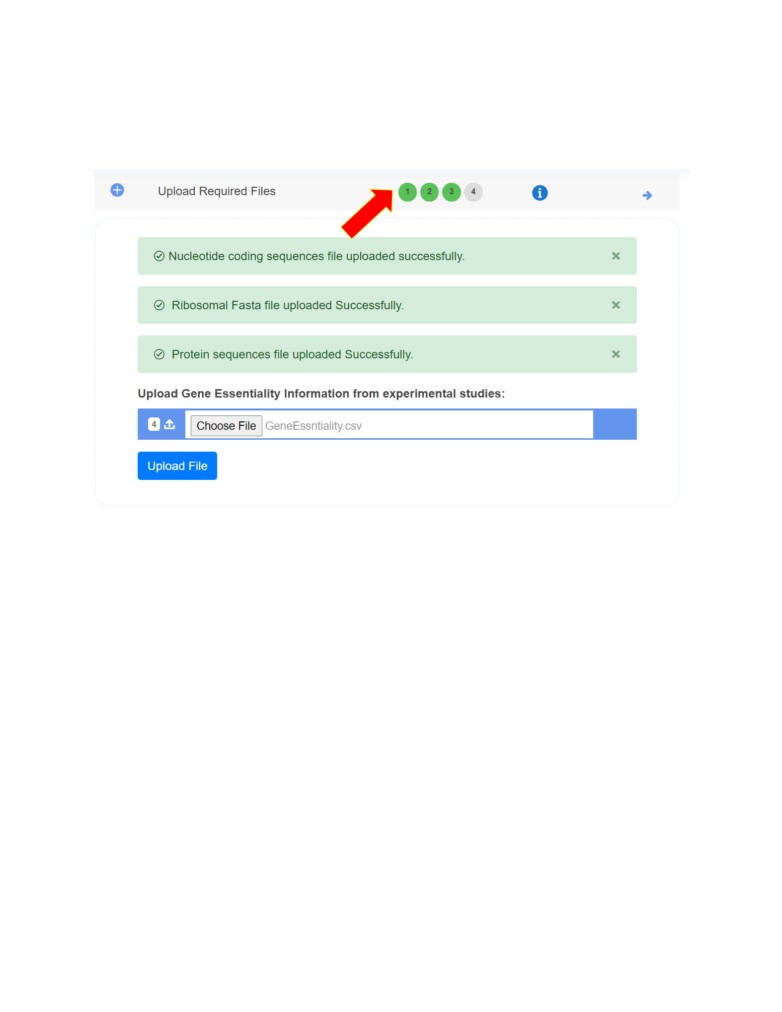
• Next step is to upload the other four required input files (see above input files 2-5).
Navigate to each next input file to upload the required category of file.
Once all the input files are uploaded successfully, you proceed to the next step in the pipeline
Dataset Preparation (Feature Matrix Calculation).
STEP 2, STEP 3 and STEP 4: repeat as Channel I
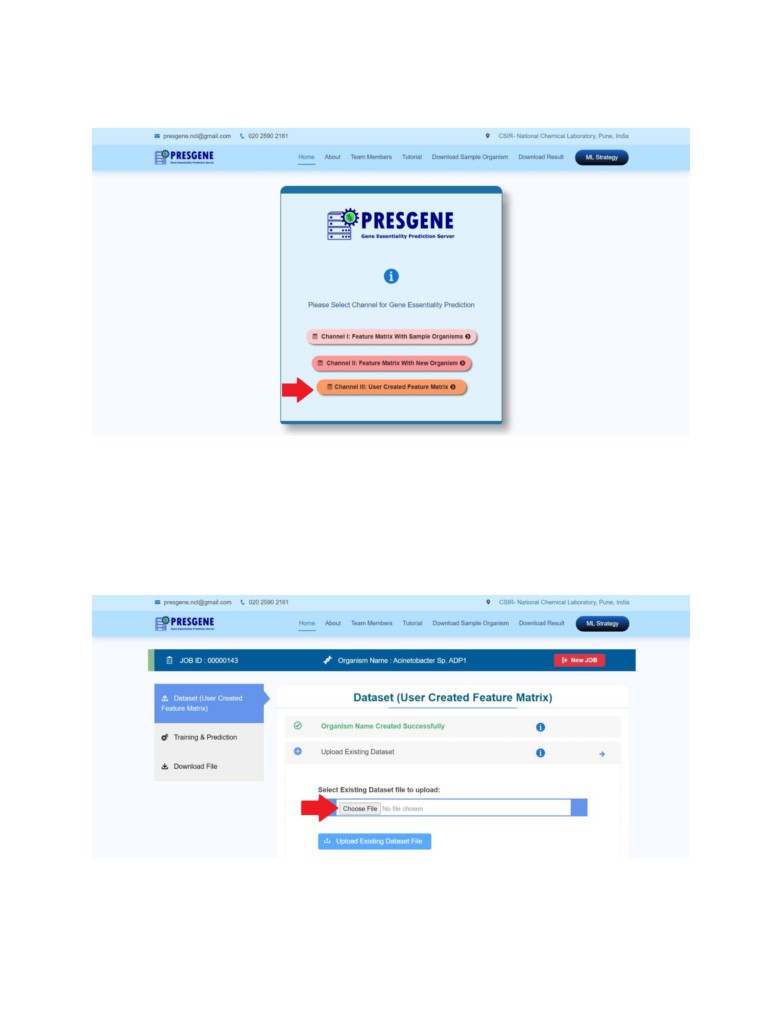
Channel III: Essential Gene prediction using user created Feature matrix
STEP 1: Enter the organism and directly proceed to upload your dataset (Feature Matrix) in .csv
format by clicking on Dataset (User Created Feature Matrix) tab on the left panel.
STEP 2: Repeat STEP 3 and 4 same as for Channel I or II.
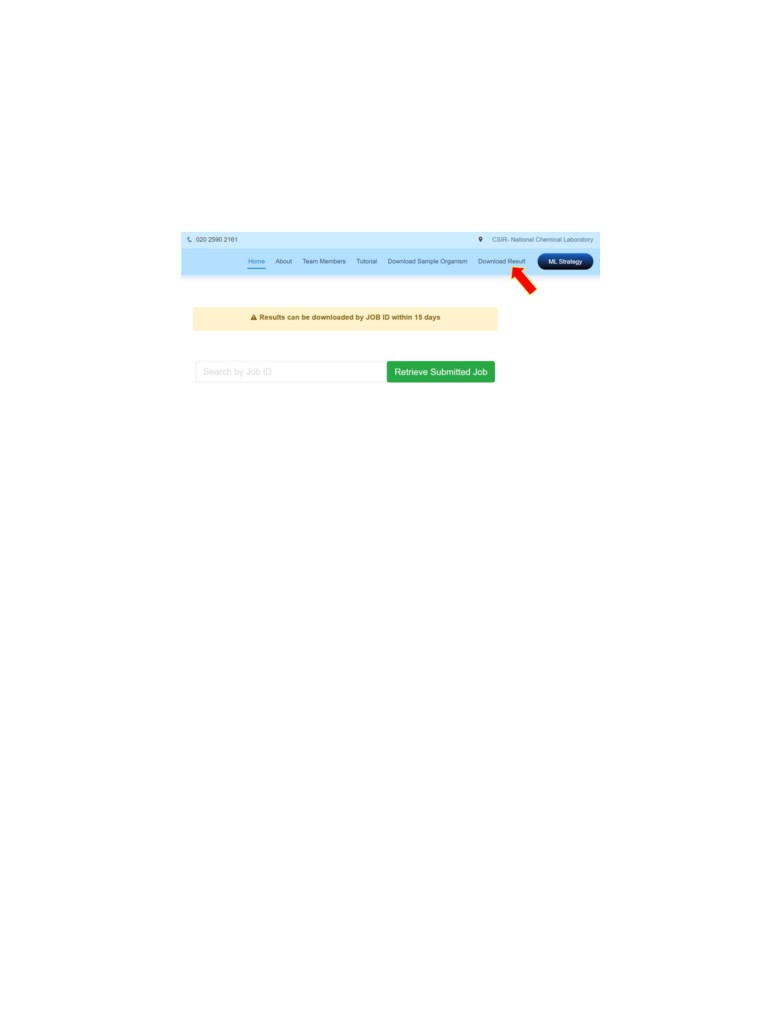
Download Results
User can click on the Download Result and enter JOB ID to retrieve previously calculated results.
These results can be retrieved within 15 days of the prediction.
References
1.
Nandi S, Subramanian A, Sarkar RR. An integrative machine learning strategy for improved
prediction of essential genes in Escherichia coli metabolism using flux-coupled features.
Mol Biosyst. 2017;13: 1584-1596. doi:10.1039/C7MB00234C
2.
Nandi S, Ganguli P, Sarkar RR. Essential gene prediction using limited gene essentiality
information-An integrative semi-supervised machine learning strategy. Mirjalili S, editor.
PLoS One. 2020;15: e0242943. doi:10.1371/journal.pone.0242943
3.
Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, et al. Ensembl 2021.
Nucleic Acids Res. 2021;49: D884-D891. doi:10.1093/nar/gkaa942
4.
Csardi G, Nepusz T, others. The igraph software package for complex network research.
5.
Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software
Suite. Trends Genet. 2000;16: 276-277. doi:10.1016/S0168-9525(00)02024-2