PRESGENE (PRediction of ESsential GENE) webserver
The minimally essential genes in an organism comprise a set of absolutely necessary genes that are required for performing vital cellular functions and are indispensable for the survival of the organism under any environmental condition. The study of essential genes in disease-causing organisms has wide application in the prediction of therapeutic targets.
Previous works from our group have demonstrated two pipelines for predicting essential genes with high accuracy that mitigates the problems of training dataset imbalance and limited availability of experimentally labeled essential genes. The first strategy (ML Strategy 1), i.e., based on the Supervised ML approach, has been developed to predict essential genes when sufficient experimental data is available, but the dataset is imbalanced (Nandi et al. Molecular BioSystems. 2017;13(8):1584-96). In the second study (ML Strategy 2), using a semi-supervised ML approach, we have proposed another pipeline for the classification of essential genes and non-essential genes of organisms where the availability of experimental data is very limited (Nandi et al., PloS one. 2020; 15.11: e0242943).
ML Strategy 1 and ML Strategy 2 based pipelines have been validated with several organisms, including both the prokaryotes and eukaryotes. These pipelines create machine learning models with high accuracy for the classification of essential and non-essential genes that addresses the problems of limited labeled data, automatic selection of heterogeneous biological features, and optimization of model parameters.
Based on these pipelines, we have developed the PRESGENE web server (Figure 1) that offers various feature calculation techniques such as network topological features of the reaction network, flux-coupled sub-network derived from the metabolic network, and sequenced-based features that are associated with gene essentiality. The list of features has been enlisted in Table 1 of the tutorial. This platform provides a user-friendly interface with integrated Machine Learning strategies that can be used to annotate essential genes in both prokaryotes and eukaryotes with high accuracy.
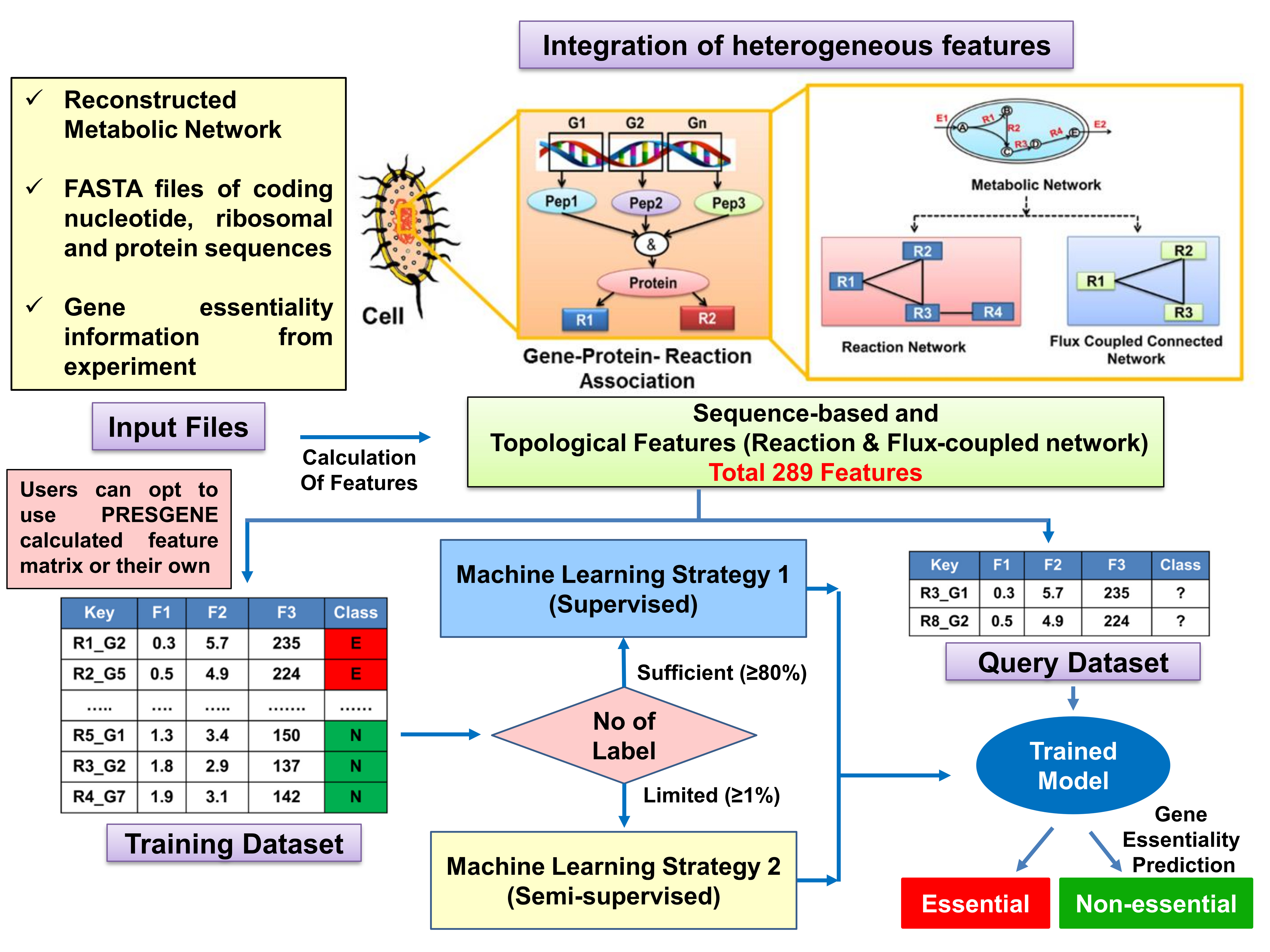
Figure 1. Workflow for PRESGENE webserver. Two pipelines are developed and integrated to predict essential genes based on sufficient unbalanced or limited labeled training dataset with sequence, informatics, topological network features derived from the genome scale reconstructed metabolic networks.
